User:Paphrag/sandbox
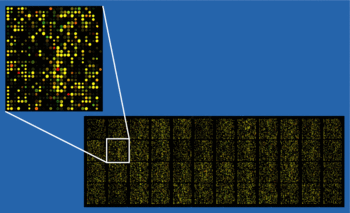
A DNA microarray (also commonly known as gene or genome chip, DNA chip, or gene array) is a collection of microscopic DNA spots attached to a solid surface, such as glass, plastic or silicon chip forming an array for the purpose of gene expression profiling, genotyping, copy number analysis, loss of heterozygosity (LOH) analysis, or DNA-protein interaction (ChIP). The principal advantage of DNA microarrays over previous technologies, is that DNA microarrays allow the biological status of thousands of genes or genomic loci to be assayed simultaneously.
The affixed DNA segments are commonly known as probes (although some sources will use different nomenclature such as reporters), thousands of which can be placed in known locations on a single DNA microarray. Microarray technology evolved from Southern blotting, whereby fragmented DNA is attached to a substrate and then probed with a known gene or fragment. Using microarrays to assay genes or genomic loci is relevant to many areas of research biology and medicine, such as understanding cell biology, biochemical pathways, developmental stages of disease, and disease treatment. For example, microarrays can be used to identify genes involved in the progression of disease by comparing gene expression profiles of diseased cells to that of normal cells.
History of DNA Microarrays
[edit]The use of microarrays for gene expression profiling was first published in 1995 (Schena et. al., 1995) (Science) and the first complete eukaryotic genome (Saccharomyces cerevisiae) on a microarray was published in 1997 (Science).
Fabrication
[edit]Microarrays are currently fabricated using a variety of technologies, including printing (spotting) with fine-pointed pins, ink-jet printing [1], photolithography, or electrochemistry.
DNA microarrays can be used to detect RNAs that may or may not be translated into active proteins. Scientists refer to this kind of analysis as "expression analysis" or expression profiling. Since there can be tens of thousands of distinct probes on an array, each microarray experiment can accomplish the equivalent number of genetic tests in parallel. Arrays have therefore dramatically accelerated many types of investigations.
Spotted microarrays
[edit]In spotted microarrays the probes, which are physically spotted onto glass slides (also occasionally plastic slides or nylon membranes), may be oligonucleotides, cDNAs, PCR products corresponding to mRNAs or other DNA probes of interest, or may consist of purified vector DNAs such as BAC DNA which contain large segments of genomic DNA. Spotted microarrays are one of the earliest published methods of producing DNA microarrays. Flexibility of probe content and low production cost make this technology attractive particularly to academic laboratories where the content of commercial arrays may not satisfy the requirements of a particular investigation, or where the higher cost of commercial arrays might be prohibitive. Spotted arrays can be subject to more technical artifacts than some other fabrication techniques due to the difficulties of quality controlling the microdynamics of liquid-solid surface interactions.
is typically hybridized with cDNA from two samples to be compared (e.g. patient and control) that are labeled with two different fluorophores (e.g. Rhodamine (red) and Fluorescein (green)). The samples can be mixed and hybridized to one single microarray that is then scanned, allowing the visualization of up-regulated and down-regulated genes in one go. The downside of this is that the absolute levels of gene expression cannot be observed, but only one chip is needed per experiment. One example of a provider for such microarrays is Eppendorf (company) with their DualChip platform.
Ink-jet Microarrays
[edit]Photolithography Microarrays
[edit]photolithography using pre-made masks, photolithography using dynamic micromirror devices
Electrochemical Microarrays
[edit]electrochemistry on microelectrode arrays.
Oligonucleotide microarrays
[edit]
In oligonucleotide microarrays (or single-channel microarrays), the probes are designed to match parts of the sequence of known or predicted mRNAs. There are commercially available designs that cover complete genomes from companies such as GE Healthcare, Affymetrix, Ocimum Biosolutions, or Agilent. These microarrays give estimations of the absolute value of gene expression and therefore the comparison of two conditions requires the use of two separate microarrays.
Oligonucleotide Arrays can be either produced by piezoelectric deposition with full length oligonucleotides or in-situ synthesis.
Long Oligonucleotide Arrays are composed of 60-mers, or 50-mers and are produced by ink-jet printing on a silica substrate. Short Oligonucleotide Arrays are composed of 25-mer or 30-mer and are produced by photolithographic synthesis (Affymetrix) on a silica substrate or piezoelectric deposition (GE Healthcare) on an acrylamide matrix. More recently, Maskless Array Synthesis from NimbleGen Systems has combined flexibility with large numbers of probes. Arrays can contain up to 390,000 spots, from a custom array design. New array formats are being developed to study specific pathways or disease states for a systems biology approach.
Oligonucleotide microarrays often contain control probes designed to hybridize with RNA spike-ins. The degree of hybridization between the spike-ins and the control probes is used to normalize the hybridization measurements for the target probes.
DNA Microarray Uses
[edit]
Genotyping microarrays
[edit]DNA microarrays can also be used to read the sequence of a genome in particular positions.
SNP microarrays are a particular type of DNA microarrays that are used to identify genetic variation in individuals and across populations. Short oligonucleotide arrays can be used to identify the single nucleotide polymorphisms (SNPs) that are thought to be responsible for genetic variation and the source of susceptibility to genetically caused diseases. Generally termed genotyping applications, DNA microarrays may be used in this fashion for forensic applications, rapidly discovering or measuring genetic predisposition to disease, or identifying DNA-based drug candidates.
These SNP microarrays are also being used to profile somatic mutations in cancer, specifically loss of heterozygosity events and amplifications and deletions of regions of DNA. Amplifications and deletions can also be detected using comparative genomic hybridization in conjunction with microarrays.
Resequencing arrays have also been developed to sequence portions of the genome in individuals. These arrays may be used to evaluate germline mutations in individuals, or somatic mutations in cancers.
Genome tiling arrays include overlapping oligonucleotides designed to blanket an entire genomic region of interest. Many companies have successfully designed tiling arrays that cover whole human chromosomes.
Microarrays and bioinformatics
[edit]
Experimental Design
[edit]Due to the biological complexity of gene expression, the considerations of experimental design that are discussed in the expression profiling article are of critical importance if statistically and biologically valid conclusions are to be drawn from the data.
Standardization
[edit]The lack of standardization in arrays presents an interoperability problem in bioinformatics, which hinders the exchange of array data. Various grass-roots open-source projects are attempting to facilitate the exchange and analysis of data produced with non-proprietary chips. The "Minimum Information About a Microarray Experiment" (MIAME) checklist helps define the level of detail that should exist and is being adopted by many journals as a requirement for the submission of papers incorporating microarray results. MIAME describes possible content but is not a format, many formats can in turn support the MIAME requirements yet there is no way to computationally determine semantic compliance.
There is currently an ongoing project being conducted by the FDA to develop standards and quality control metrics which will eventually allow the use of MicroArray data in drug discovery, clinical practice and regulatory decision-making. Detailed information about the FDA's MicroArray Quality Control (MAQC) Project is available at http://www.fda.gov/nctr/science/centers/toxicoinformatics/maqc/ . The MicroArray and Gene Expression (MAGE) group is working on the standardization of the representation of gene expression data and relevant annotations.
Statistical analysis
[edit]The analysis of DNA microarrays poses a large number of statistical problems, including the normalization of the data. There are dozens of proposed normalization methods in the published literature; as in many other cases where authorities disagree, a sound conservative approach is to try a number of popular normalization methods and compare the conclusions reached: how sensitive are the main conclusions to the method chosen?
From a hypothesis-testing perspective, the large number of genes present on a single array means that the experimenter must take into account a multiple testing problem: even if each gene is extremely unlikely to randomly yield a result of interest, the combination of all the genes is likely to show at least one or a few occurrences of this result which are false positives.
A basic difference between microarray data analysis and much traditional biomedical research is the dimensionality of the data. A large clinical study might collect 100 data items per patient for thousands of patients. A medium-size microarray study will obtain many thousands of numbers per sample for perhaps a hundred samples. Many analysis techniques treat each sample as a single point in a space with thousands of dimensions, then attempt by various techniques to reduce the dimensionality of the data to something humans can visualize.
Relation between probe and gene
[edit]The relation between a probe and the mRNA that it is expected to detect is problematic. On the one hand, some mRNAs may cross-hybridize probes in the array that are supposed to detect another mRNA. On the other hand, probes that are designed to detect the mRNA of a particular gene may be relying on genomic EST information that is incorrectly associated with that gene.
Public databases to analyse microarray data
[edit]- Stanford Microarray database
- Yale Microarray Database
- UNC Microarray database
- MUSC database
- Gene Expression Omnibus - NCBI
- ArrayExpress - EBI
- University of Tennessee Microarray Database
- List of a nearly 40 further microarray databases
List of notable microarray technology companies
[edit]- Affymetrix
- Agilent Technologies
- Asper Biotech
- CombiMatrix
- Eppendorf
- febit
- GE Healthcare
- GenePix
- Illumina
- Nanogen
- Nimblegen Systems
- Ocimum Biosolutions (acquired from MWG Biotech)
- Roche Diagnostics
External links
[edit]{{cleanup-spam}}
- DNA Microarrays in Health Care and Drug Discovery
- PLoS Biology Primer: Microarray Analysis
- Leming Shi's Genome Chip Resources Non-commercial site with references
- Nature Genetics Free Issue on Gene Chips
- DNA Microarray Methodology (A brilliant flash movie)
- How to build your own arrayer (Stanford University)
- How to build your own ink jet microarrayer article in Genome Biology (open access)
- Microarray protocols, how-to documents, free software
- Microarrayer tools and resources
- Rundown of microarray technology
- Large-Scale Gene Expression and Microarray Links and Resources
- Microarray data analysis
- Microarray Data Classification Server (MDCS)
- TiMAT: Tiling Microarray Analysis Tools
- The Microarray Gene Expression Data Society, and home of MIAME
- Try DNA microarray yourself in an interactive demonstration
- ArrayExpress at the European Bioinformatics Institute
- Gene Expression Omnibus (GEO) at NCBI
- Genevestigator at ETH Zurich
- Center for Functional Genomics, SUNY Albany's core lab providing microarray and related services to all
- The Science Creative Quarterly's overview of Microarrays - also excellent free hi-res schematic images available on the technique itself.
References
[edit]Schena M, Shalon D, Davis RW, Brown PO. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. Oct 20; 270 (5235): 467-70.
