User:MochiBee/RBFOX1
| This is not a Wikipedia article: It is an individual user's work-in-progress page, and may be incomplete and/or unreliable. For guidance on developing this draft, see Wikipedia:So you made a userspace draft. Find sources: Google (books · news · scholar · free images · WP refs) · FENS · JSTOR · TWL |
| RBFOX1 | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||
| Aliases | RBFOX1, 2BP1, A2BP1, FOX-1, FOX1, HRNBP1, RNA binding protein, fox-1 homolog 1, RNA binding fox-1 homolog 1 | ||||||||||||||||||||||||||||||
| External IDs | OMIM: 605104; MGI: 1926224; HomoloGene: 69339; GeneCards: RBFOX1; OMA:RBFOX1 - orthologs | ||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
Fox-1 homolog A, also known as ataxin 2-binding protein 1 (A2BP1) or hexaribonucleotide-binding protein 1 (HRNBP1) or RNA binding protein, fox-1 homolog (Rbfox1), is a protein that in humans is encoded by the RBFOX1 gene.
Discovery
[edit]The RBFOX1 gene was first studied in Caenorhabditis elegans (nematodes), Drosophila melanogaster (fruit flies), and Danio rerio (zebrafish) with origins in embryology and development. The derivation of the nomenclature for RBFOX1 comes from the original sexual differentiation studies in C. elegans where the gene was denoted as 'Feminizing locus On X' (Fox-1). This refers to a lethal splicing event which causes an increase in the chromosomal X:A ratio; feminizing XO males. In Drosophila, the gene is known as CG3206 and was noted to code for an RNA-binding protein, be affected by Notch-signaling, and be associated with non-D/V (dorso-ventral) cells of the wing discs during wing development. The 'RB' portion of the gene's name extends from the RNA-binding (RB) properties of the coded protein. In zebrafish, rbfox genes were identified as being essential for cardiac and skeletal muscle development, causing reduced heart rate and paralysis respectively in morphants. The discovery of RBFOX1 in humans was due to the interaction of Rbfox1 with ataxin-2, hence the alternative name of A2BP1 (or ataxin-2 binding protein-1).[5]
Structure
[edit]RBFOX1 is located on chromosome 16 and consists of 30 exons. The Rbfox1 protein consists of 397 amino acids (AA) and is 42,784 Da. The canonical folding of the protein includes three beta sheets and two alpha helices. The localization of Rbfox1 protein is determined by its own alternative splicing via RBFOX proteins. If exon 19 is included, Rbfox1 will be cytoplasmic, but if exon 19 is excluded, Rbfox1 will be nuclear.
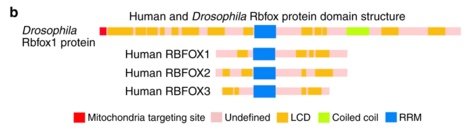
There are five isoforms of Rbfox1 due to alternative splicing. The canonical variant, isoform 1, is also known as gamma. This RBFOX1 transcript includes three conserved domains in its sequence. The most clinically relevant of these domains is the RNA recognition motif (RRM) located between 137-212. This domain allows for the important property of RNA binding for the Rbfox1 protein. Another conserved domain of RBFOX1 is the calcitonin gene-related peptide regulator C terminal. RBFOX1's C terminal is located between 273-363 and, as the name suggests, regulates the calcitonin gene-related peptide. The third conserved domain of RBFOX1 is the ELAV/HuD family splicing factor. HUD is human paraneoplastic encephalomyelitis antigen D whereas ELAV is Drosophila embryonic lethal abnormal visual protein. ELAV-like splicing factors are also known in humans as HuB (ELAV-like protein 2), HuC (ELAV-like protein 3, Paraneoplastic cerebellar degeneration-associated antigen), and HuR (ELAV-like protein 1). This super family domain contains three RRMs and is located between 25-208.[7]
| SNP | Alleles | AA Change | Type | Location | Associated Disease | Ref | |
|---|---|---|---|---|---|---|---|
| Common | rs147023054 | C>T | intron variation | [8] | |||
| rs372761949 | G/A | V180M | missense variation | [8] | |||
| rs974157467 | ACTGCCG/A | inframe deletion | [8] | ||||
| rs145873257 | G/A | G353S | missense variation | [8] | |||
| rs2093621567 | CA/C | frameshift variation | [8] | ||||
| Disease Relevant | rs12921846 | A>T | intron variation | intron 3 | Conduct Disorder in ADHD | [8][9] | |
| rs10153149 | A>C | intron variation | intron 3 | Conduct Disorder in ADHD | [9] | ||
| rs9940753 | G>C | intron variation | ADHD, ASD | [8] | |||
| rs12447542 | Unknown | Schizophrenia | [9] | ||||
| rs12444931 | G>A | intron variation | Schizophrenia, Bipolar Disorder | [8][9] | |||
| rs133341055 | T>G | intron variation | intron 1 | Anxiety | [9] | ||
| rs809682 | Unknown | Anxiety | [9] | ||||
| rs142723691 | A>G | intron variation | Hepatitis A | [8] | |||
| rs6500818 | C>T | intron variation | Dengue Shock Syndrome | [8] | |||
| rs192187627 | A>C | intron variation | COVID-19 | [8] |
There are forty possible isoforms but only five beyond the canonical sequence are understood and confirmed in the population. Isoform 2 of RBFOX1, also known as alpha, is a shorter form of the canonical sequence as it is missing an in-frame segment on the 3' coding region. The third variant, beta, is also a shorter version of isoform 1. This shortening is caused by an alternate exon in the 3' coding region. Because of this, isoform 3 has a differing C-terminus located between 273-360. RBFOX1's isoform 4 differs in that the 5'UTR (untranslated region) lacks an in-frame section of the 3' coding region. This shorter isoform is encoded by variants 4 and 6 and has an alternate N-terminus. This isoform includes changes of locations of two of the conserved domains and one other domain: cell division protein ZipA becomes located between 4-122, calcitonin gene-related peptide regulator C terminal becomes located between 253-342, and the RNA recognition motif becomes located between 117-192. Isoform 5 contains a different 5'UTR as well as multiple coding region differences. Beyond these internal differences, isoform 5 also has a shorter and distinct N-terminus. The C terminus is located between 226-315 while the RRM domain is located between 117-192. The ZipA protein domain is located between 4-122. The differences of isoform 6 results in the use of an alternate start codon and a frameshift in the 3' coding region. The UTR is changed and multiple coding regions are altered. Uniquely, this isoform contains a longer rather than shorter N-terminus and a distinct C-terminus. The locations for the ZipA protein, calcitonin gene-related peptide regulator C terminal, and RRM are 33-165, 296-385, and 160-235, respectively.[8]
Function
[edit]
RBFOX1 is expressed in human heart, muscle, and neuronal tissues. The primary function is regulation of alternative splicing of associated genes. Several alternatively spliced transcript variants have been found for this gene with some localizing to the nucleus and others to the cytoplasm. Nuclear variants have a well-established role in tissue specific alternative splicing. Rbfox1 cytoplasmic variants modulate mRNA stability and translation. In stressed cells, Rbfox1 has been demonstrated to localize to cytoplasmic stress granules. Rbfox1 has an RNA recognition motif that is highly conserved among RNA-binding proteins. Rbfox1, and the related protein Rbfox2, bind the consensus RNA sequence motif (U)GCAUG within introns to exert their functions as alternative splicing factors. The C terminus of RBFOX1 contains the code for a protein, calcitonin gene-related peptide, involved with mediating neuron-specific splicing. Together, Rbfox1 and Rbfox2 repress exon 4 inclusion. In particular, for Drosophila, two cytoplasmic Rbfox1 isoforms bind Pumilio mRNA for silencing. Because of this destabilization, germline development is promoted and reversion to earlier stages is prevented.[11][12] The alternative splicing activity of RBFOX1 also aids in neuronal development specifically for CaV1.2 voltage-gated calcium channels and N-methyl-D-aspartate (NMDA) receptors.[13][14] The overall activity and molecular mechanism of alternative splicing mediation for RBFOX1 is not fully understood, but some qualities have been established in recent studies. For example, exon inclusion is sufficiently promoted with only the carboxy terminal tethered downstream of the alternative exon. Conversely, for repression, both the RNA binding motif and carboxy terminal are required when tethered upstream of the alternative exon. Possible proteins that aid in the inclusion or skipping process are not confirmed, though both hnRNPH and RALY have been shown to bind Rbfox1. Thus, the specific mechanisms of alternative splicing maintenance via RBFOX1 are unknown. In one study, dominant-negative RBFOX protein interfered with exon activation, though not exon skipping. Because of this knowledge, repression maintenance most likely includes other proteins or outside factors near the binding sites. In C. elegans, co-operative binding between SUP-12 and RBFOX1 is noted to account for tissue-specific splicing. In mammals, there is a more universal cooperativity between RBFOX and NOVA family of proteins. The overall repression and inclusion activity of exons via RBFOX1 seem to be positionally-related. That is, a location downstream of an intron would lead to exon inclusion and a location upstream of an intron would lead to exon exclusion.[15][5][16]
Neurodevelopmental Disorders (NDDs)
[edit]Autism Spectrum Disorder (ASD)
[edit]ASD is a neurodevelopmental disorder of social communication and repetitive behaviors as well as fixated interests and/or sensory behavior. Typically, ASD is diagnosed in adolescence, but it is possible to be diagnosed in later stages of life. According to the DSM-5-TR, an ASD diagnosis requires at least two of the four restricted repetitive behaviors and all three verbal or nonverbal communication deficits.[17][18] Mutations of RBFOX1 are not sufficient to single-handedly develop autism, but rather also require an environmental risk factor. Numerous ASD samples from cohorts and isolated autistic patients have been linked to de novo copy number variations (CNVs) of RBFOX1. Universally, cases from these studies involved intragenic deletions of either exons 5, 6, or 1D. In human progenitor cell lines (a stem cell culture method) modeling haploinsufficiency in neuronal differentiation, a knockdown (interference with gene or protein activity) study of RBFOX1 revealed significant changes in RNA splicing and gene expression. Similarly, whole transcriptome analysis of ASD patients showcased a reduction of RBFOX1 and dysregulation of RBFOX1-dependent alternative splicing.[5][9] RBFOX1 also contributes to mRNA stability of autism-related genes by blocking miRNA binding.[12]

Epilepsy
[edit]While epilepsy, episodes of recurrent seizures, is most notably a neurological disorder, there are some cases which link the disease to issues with neuronal development. The two types of seizures are convulsive (60%) and non-convulsive (40%) with varying subcategories in each branch. A seizure is sporadic neural activity with no purpose.[19][20] Interestingly, there is some comorbidity between ASD and epilepsy. Though it is unknown the specifics of how RBFOX1 affects neuronal development, it has been shown in neural-specific mouse knockouts that synaptic transmission and increased membrane excitability occur, causing a predisposition to seizures.[5][21][22] RBFOX1 potentially provides mRNA stability for synaptic genes by blocking miRNA binding.[12]
Attention Deficit Hyperactivity Disorder (ADHD)
[edit]While it is not agreed upon the causes of ADHD, it is known there are genetic risk factors that can contribute to the predisposition to the disorder. Oftentimes, a diagnosis requires a series of tests, observations, and questionnaires with the patient proving at least six of the nine inattentive and at least six of the nine hyperactivity and impulsivity symptoms (according to the DSM-5).[23] Because RBFOX1 has been noted to affect neuronal migration and synapse formation, there may be reasonable concern for its contribution to predisposition of ADHD.[24]
Schizophrenia
[edit]Schizophrenia is a disorder with both positive (delusions, hallucinations, and disorganized thought) and negative (povery of speech, social withdrawal, and flattened effect) symptoms. In some individual studies, copy number variations of RBFOX1 have been linked with schizophrenia at low levels with a notable increase in risk for male-specific schizophrenia. This increased risk is said to be due to a duplication before exon 6.[12]
Neurodegenerative Diseases
[edit]
Spinocerebellar Ataxia (SCA)
[edit]SCA is a neurodegenerative disease that slowly impedes gait, causes slurred speech, and causes an inability to control motor functions such as balance and coordination. This group of ataxias typically do not begin until adulthood. Several mechanisms play into the manifestation of this disease including ion channel dysfunction, RNA toxicity, and proteotoxicity. Due to the heterogenous nature of SCA, therapies are very difficult to develop and would most likely require specificity for each type. Rbfox1 is noted to be a possible contributor to spinocerebellar ataxia type 2 (SCA2), one of twelve dominant repeat expansion SCAs. The repeat is a CAG and causes an excessive string of glutamines to be translated. Unlike most other SCAs which are purely cerebral, SCA2 also includes neurodegeneration. The Rbfox1/A2BP1 protein binds to the C-terminus of ataxin-2, and may contribute to the restricted pathology of SCA2. Ataxin-2 is the gene product of the SCA2 gene which causes familial diseases. The polyglutamine SCAs not only have RNA foci and proteinaceous inclusions, but also the misfolded proteins themselves seem to aggregate in neuronal nuclei.[25]

Alzheimer's Disease (AD)
[edit]AD is a complex disease with different contributing pathological aspects. The most agreed upon pathologies are amyloid plaques, neurofibrillary tau tangles, and neuroinflammation. The amyloid plaques are extracellular with respect to neurons and occur early on in neurodegenerative diseases. Tau aids in the intracellular structure of the neuron by binding to and strengthening microtubules. When mutated, the tau can abnormally phosphorylate or misfold and bind to itself, causing tangles that damage the neuron. These tangles are typically seen in the later stages of neurodegenerative diseases.[27] In healthy humans, misfolded tau can be cleared from the system by the ubiquitin-proteasome system (UPS) or the autophagy-lysosome pathway.[28] In genetically predisposed or aged humans, these systems lose efficiency and can no longer handle the accumulating amount of misfolded tau, causing tangles to form more often without a way of clearing. One aspect of predisposition includes different isoforms of the beta amyloid precursor protein (APP). These isoforms are caused by varying cleavages of APP by either beta secretase and gamma secretase or alpha secretase. The longer forms of APP are prone to aggregating and causing disruptions of the system.[29] In particular, within in vitro experiments, RBFOX1 upregulation seems to be associated with an increase in the APP714 isoform. This isoform excludes exon 7 without including exon 8 of the APP, causing a shorter form of APP. In AD patient brains, RBFOX1 was downregulated in the dorsolateral prefrontal cortex tissue; this points to the possibility of RBFOX1 playing a role in alternative splicing within the prefrontal cortex and contributing to control of plaques.[30] With regards to neuroinflammatory contribution to AD, RBFOX1 also may have ties with microglia. According to genome wide association (GWA) data, moduleQTL (modQTL) RBFOX1 SNP may alter gene expression of microglia.[31][9]
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000078328 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000008658 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c d Bill, Brent R.; Lowe, Jennifer K.; DyBuncio, Christina T.; Fogel, Brent L. (2013), "Orchestration of Neurodevelopmental Programs by RBFOX1", International Review of Neurobiology, vol. 113, Elsevier, pp. 251–267, doi:10.1016/b978-0-12-418700-9.00008-3, ISBN 978-0-12-418700-9, PMC 4318517, PMID 24290388, retrieved 2023-11-03
{{citation}}: CS1 maint: PMC format (link) - ^ Kucherenko, Mariya M.; Shcherbata, Halyna R. (2018-01-22). "Stress-dependent miR-980 regulation of Rbfox1/A2bp1 promotes ribonucleoprotein granule formation and cell survival". Nature Communications. 9 (1): 312. doi:10.1038/s41467-017-02757-w. ISSN 2041-1723. PMC 5778076. PMID 29358748.
{{cite journal}}: CS1 maint: PMC format (link) - ^ "NCBI Conserved Domain Search". www.ncbi.nlm.nih.gov. Retrieved 2023-11-29.
- ^ a b c d e f g h i j k l "RBFOX1 RNA binding fox-1 homolog 1 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2023-11-29.
- ^ a b c d e f g h Fernàndez-Castillo, Noèlia; Gan, Gabriela; van Donkelaar, Marjolein M. J.; Vaht, Mariliis; Weber, Heike; Retz, Wolfgang; Meyer-Lindenberg, Andreas; Franke, Barbara; Harro, Jaanus; Reif, Andreas; Faraone, Stephen V.; Cormand, Bru (2020-01-01). "RBFOX1, encoding a splicing regulator, is a candidate gene for aggressive behavior". European Neuropsychopharmacology. Neurobiology of aggressive behavior in the context of ADHD and related disorders. 30: 44–55. doi:10.1016/j.euroneuro.2017.11.012. ISSN 0924-977X.
- ^ "26 items (human) - STRING interaction network". string-db.org. Retrieved 2023-12-04.
- ^ Carreira-Rosario, Arnaldo; Bhargava, Varsha; Hillebrand, Jens; Kollipara, Rahul K.; Ramaswami, Mani; Buszczak, Michael (2016-03). "Repression of Pumilio Protein Expression by Rbfox1 Promotes Germ Cell Differentiation". Developmental Cell. 36 (5): 562–571. doi:10.1016/j.devcel.2016.02.010. ISSN 1534-5807. PMC 4785839. PMID 26954550.
{{cite journal}}: Check date values in:|date=(help); no-break space character in|first4=at position 6 (help)CS1 maint: PMC format (link) - ^ a b c d Baralle, Francisco E.; Giudice, Jimena (2017-07). "Alternative splicing as a regulator of development and tissue identity". Nature Reviews Molecular Cell Biology. 18 (7): 437–451. doi:10.1038/nrm.2017.27. ISSN 1471-0080. PMC 6839889. PMID 28488700.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Keller, Roberto; Basta, Roberta; Salerno, Luana; Elia, Maurizio (2017-08-01). "Autism, epilepsy, and synaptopathies: a not rare association". Neurological Sciences. 38 (8): 1353–1361. doi:10.1007/s10072-017-2974-x. ISSN 1590-3478.
- ^ Fisher, Emily; Feng, Jian (2022-11). "RNA splicing regulators play critical roles in neurogenesis". WIREs RNA. 13 (6). doi:10.1002/wrna.1728. ISSN 1757-7004.
{{cite journal}}: Check date values in:|date=(help) - ^ Sun, Shuying; Zhang, Zuo; Fregoso, Oliver; Krainer, Adrian R. (2012-02-01). "Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2". RNA. 18 (2): 274–283. doi:10.1261/rna.030486.111. ISSN 1355-8382. PMID 22184459.
- ^ Fu, Xiang-Dong; Ares, Manuel (2014-10). "Context-dependent control of alternative splicing by RNA-binding proteins". Nature Reviews Genetics. 15 (10): 689–701. doi:10.1038/nrg3778. ISSN 1471-0064. PMC 4440546. PMID 25112293.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ First, Michael B.; Yousif, Lamyaa H.; Clarke, Diana E.; Wang, Philip S.; Gogtay, Nitin; Appelbaum, Paul S. (2022-06). "DSM‐5‐TR: overview of what's new and what's changed". World Psychiatry. 21 (2): 218–219. doi:10.1002/wps.20989. ISSN 1723-8617. PMC 9077590. PMID 35524596.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Lord, Catherine; Brugha, Traolach S.; Charman, Tony; Cusack, James; Dumas, Guillaume; Frazier, Thomas; Jones, Emily J. H.; Jones, Rebecca M.; Pickles, Andrew; State, Matthew W.; Taylor, Julie Lounds; Veenstra-VanderWeele, Jeremy (2020-01-16). "Autism spectrum disorder". Nature Reviews Disease Primers. 6 (1): 1–23. doi:10.1038/s41572-019-0138-4. ISSN 2056-676X. PMC 8900942. PMID 31949163.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Bozzi, Yuri; Casarosa, Simona; Caleo, Matteo (2012). "Epilepsy as a Neurodevelopmental Disorder". Frontiers in Psychiatry. 3. doi:10.3389/fpsyt.2012.00019. ISSN 1664-0640. PMC 3306997. PMID 22457654.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Beghi, Ettore (2020). "The Epidemiology of Epilepsy". Neuroepidemiology. 54 (2): 185–191. doi:10.1159/000503831. ISSN 0251-5350.
- ^ Norris, Adam; Calarco, John (2012). "Emerging Roles of Alternative Pre-mRNA Splicing Regulation in Neuronal Development and Function". Frontiers in Neuroscience. 6. doi:10.3389/fnins.2012.00122. ISSN 1662-453X. PMC 3424503. PMID 22936897.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Bryant, C. D.; Yazdani, N. (2016-01). "RNA ‐binding proteins, neural development and the addictions". Genes, Brain and Behavior. 15 (1): 169–186. doi:10.1111/gbb.12273. ISSN 1601-1848. PMC 4944654. PMID 26643147.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Thapar, Anita; Cooper, Miriam; Eyre, Olga; Langley, Kate (2013-01). "Practitioner Review: What have we learnt about the causes of ADHD?". Journal of Child Psychology and Psychiatry. 54 (1): 3–16. doi:10.1111/j.1469-7610.2012.02611.x. ISSN 0021-9630. PMC 3572580. PMID 22963644.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Ribasés, M.; Mitjans, M.; Hartman, CA; Soler Artigas, M.; Demontis, D.; Larsson, H.; Ramos-Quiroga, JA; Kuntsi, J.; Faraone, SV; Børglum, AD; Reif, A.; Franke, B.; Cormand, B. (2023-10-01). "Genetic architecture of ADHD and overlap with other psychiatric disorders and cognition-related phenotypes". Neuroscience & Biobehavioral Reviews. 153: 105313. doi:10.1016/j.neubiorev.2023.105313. ISSN 0149-7634.
- ^ Klockgether, Thomas; Mariotti, Caterina; Paulson, Henry L. (2019-04-11). "Spinocerebellar ataxia". Nature Reviews Disease Primers. 5 (1): 1–21. doi:10.1038/s41572-019-0074-3. ISSN 2056-676X.
- ^ "What Happens to the Brain in Alzheimer's Disease?". National Institute on Aging. Retrieved 2023-12-04.
- ^ Mandelkow, Eva-Maria; Mandelkow, Eckhard (1998-11-01). "Tau in Alzheimer's disease". Trends in Cell Biology. 8 (11): 425–427. doi:10.1016/S0962-8924(98)01368-3. ISSN 0962-8924.
- ^ Gorantla, Nalini Vijay; Chinnathambi, Subashchandrabose (2021-08-01). "Autophagic Pathways to Clear the Tau Aggregates in Alzheimer's Disease". Cellular and Molecular Neurobiology. 41 (6): 1175–1181. doi:10.1007/s10571-020-00897-0. ISSN 1573-6830.
- ^ Zhang, Yun-wu; Thompson, Robert; Zhang, Han; Xu, Huaxi (2011-01-07). "APP processing in Alzheimer's disease". Molecular Brain. 4 (1): 3. doi:10.1186/1756-6606-4-3. ISSN 1756-6606. PMC 3022812. PMID 21214928.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Nikom, David; Zheng, Sika (2023-08). "Alternative splicing in neurodegenerative disease and the promise of RNA therapies". Nature Reviews Neuroscience. 24 (8): 457–473. doi:10.1038/s41583-023-00717-6. ISSN 1471-0048.
{{cite journal}}: Check date values in:|date=(help) - ^ McFarland, Karen N.; Chakrabarty, Paramita (2022-01-01). "Microglia in Alzheimer's Disease: a Key Player in the Transition Between Homeostasis and Pathogenesis". Neurotherapeutics. 19 (1): 186–208. doi:10.1007/s13311-021-01179-3. ISSN 1878-7479. PMC 9130399. PMID 35286658.
{{cite journal}}: CS1 maint: PMC format (link)






