Nucleotide
| Part of a series on |
| Genetics |
|---|
 |
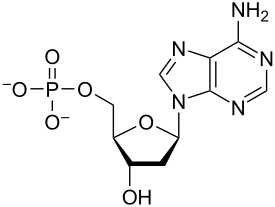
Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.[1]
Nucleotides are composed of three subunit molecules: a nucleobase, a five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine, and thymine; in RNA, uracil is used in place of thymine.
Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), and uridine triphosphate (UTP)—throughout the cell for the many cellular functions that demand energy, including: amino acid, protein and cell membrane synthesis, moving the cell and cell parts (both internally and intercellularly), cell division, etc..[2] In addition, nucleotides participate in cell signaling (cyclic guanosine monophosphate or cGMP and cyclic adenosine monophosphate or cAMP) and are incorporated into important cofactors of enzymatic reactions (e.g., coenzyme A, FAD, FMN, NAD, and NADP+).
In experimental biochemistry, nucleotides can be radiolabeled using radionuclides to yield radionucleotides.
5-nucleotides are also used in flavour enhancers as food additive to enhance the umami taste, often in the form of a yeast extract.[3]
Structure
[edit]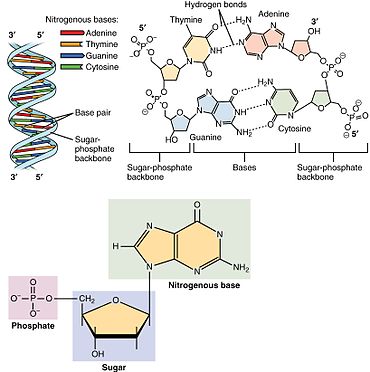
A nucleotide is composed of three distinctive chemical sub-units: a five-carbon sugar molecule, a nucleobase (the two of which together are called a nucleoside), and one phosphate group. With all three joined, a nucleotide is also termed a "nucleoside monophosphate", "nucleoside diphosphate" or "nucleoside triphosphate", depending on how many phosphates make up the phosphate group.[4]
In nucleic acids, nucleotides contain either a purine or a pyrimidine base—i.e., the nucleobase molecule, also known as a nitrogenous base—and are termed ribonucleotides if the sugar is ribose, or deoxyribonucleotides if the sugar is deoxyribose. Individual phosphate molecules repetitively connect the sugar-ring molecules in two adjacent nucleotide monomers, thereby connecting the nucleotide monomers of a nucleic acid end-to-end into a long chain. These chain-joins of sugar and phosphate molecules create a 'backbone' strand for a single- or double helix. In any one strand, the chemical orientation (directionality) of the chain-joins runs from the 5'-end to the 3'-end (read: 5 prime-end to 3 prime-end)—referring to the five carbon sites on sugar molecules in adjacent nucleotides. In a double helix, the two strands are oriented in opposite directions, which permits base pairing and complementarity between the base-pairs, all which is essential for replicating or transcribing the encoded information found in DNA.[citation needed]
Nucleic acids then are polymeric macromolecules assembled from nucleotides, the monomer-units of nucleic acids. The purine bases adenine and guanine and pyrimidine base cytosine occur in both DNA and RNA, while the pyrimidine bases thymine (in DNA) and uracil (in RNA) occur in just one. Adenine forms a base pair with thymine with two hydrogen bonds, while guanine pairs with cytosine with three hydrogen bonds.
In addition to being building blocks for the construction of nucleic acid polymers, singular nucleotides play roles in cellular energy storage and provision, cellular signaling, as a source of phosphate groups used to modulate the activity of proteins and other signaling molecules, and as enzymatic cofactors, often carrying out redox reactions. Signaling cyclic nucleotides are formed by binding the phosphate group twice to the same sugar molecule, bridging the 5'- and 3'- hydroxyl groups of the sugar.[2] Some signaling nucleotides differ from the standard single-phosphate group configuration, in having multiple phosphate groups attached to different positions on the sugar.[5] Nucleotide cofactors include a wider range of chemical groups attached to the sugar via the glycosidic bond, including nicotinamide and flavin, and in the latter case, the ribose sugar is linear rather than forming the ring seen in other nucleotides.

Synthesis
[edit]Nucleotides can be synthesized by a variety of means, both in vitro and in vivo.[citation needed]
In vitro, protecting groups may be used during laboratory production of nucleotides. A purified nucleoside is protected to create a phosphoramidite, which can then be used to obtain analogues not found in nature and/or to synthesize an oligonucleotide.[citation needed]
In vivo, nucleotides can be synthesized de novo or recycled through salvage pathways.[1] The components used in de novo nucleotide synthesis are derived from biosynthetic precursors of carbohydrate and amino acid metabolism, and from ammonia and carbon dioxide. Recently it has been also demonstrated that cellular bicarbonate metabolism can be regulated by mTORC1 signaling.[6] The liver is the major organ of de novo synthesis of all four nucleotides. De novo synthesis of pyrimidines and purines follows two different pathways. Pyrimidines are synthesized first from aspartate and carbamoyl-phosphate in the cytoplasm to the common precursor ring structure orotic acid, onto which a phosphorylated ribosyl unit is covalently linked. Purines, however, are first synthesized from the sugar template onto which the ring synthesis occurs. For reference, the syntheses of the purine and pyrimidine nucleotides are carried out by several enzymes in the cytoplasm of the cell, not within a specific organelle. Nucleotides undergo breakdown such that useful parts can be reused in synthesis reactions to create new nucleotides.[citation needed]
Pyrimidine ribonucleotide synthesis
[edit]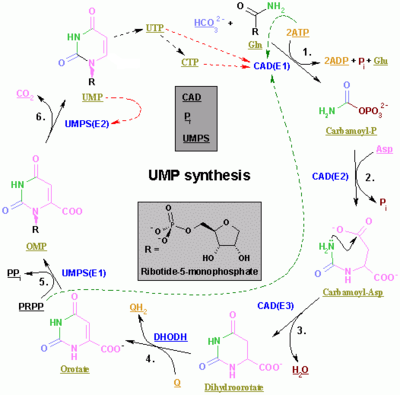
The synthesis of the pyrimidines CTP and UTP occurs in the cytoplasm and starts with the formation of carbamoyl phosphate from glutamine and CO2. Next, aspartate carbamoyltransferase catalyzes a condensation reaction between aspartate and carbamoyl phosphate to form carbamoyl aspartic acid, which is cyclized into 4,5-dihydroorotic acid by dihydroorotase. The latter is converted to orotate by dihydroorotate oxidase. The net reaction is:
- (S)-Dihydroorotate + O2 → Orotate + H2O2
Orotate is covalently linked with a phosphorylated ribosyl unit. The covalent linkage between the ribose and pyrimidine occurs at position C1[7] of the ribose unit, which contains a pyrophosphate, and N1 of the pyrimidine ring. Orotate phosphoribosyltransferase (PRPP transferase) catalyzes the net reaction yielding orotidine monophosphate (OMP):
- Orotate + 5-Phospho-α-D-ribose 1-diphosphate (PRPP) → Orotidine 5'-phosphate + Pyrophosphate
Orotidine 5'-monophosphate is decarboxylated by orotidine-5'-phosphate decarboxylase to form uridine monophosphate (UMP). PRPP transferase catalyzes both the ribosylation and decarboxylation reactions, forming UMP from orotic acid in the presence of PRPP. It is from UMP that other pyrimidine nucleotides are derived. UMP is phosphorylated by two kinases to uridine triphosphate (UTP) via two sequential reactions with ATP. First, the diphosphate from UDP is produced, which in turn is phosphorylated to UTP. Both steps are fueled by ATP hydrolysis:
- ATP + UMP → ADP + UDP
- UDP + ATP → UTP + ADP
CTP is subsequently formed by the amination of UTP by the catalytic activity of CTP synthetase. Glutamine is the NH3 donor and the reaction is fueled by ATP hydrolysis, too:
- UTP + Glutamine + ATP + H2O → CTP + ADP + Pi
Cytidine monophosphate (CMP) is derived from cytidine triphosphate (CTP) with subsequent loss of two phosphates.[8][9]
Purine ribonucleotide synthesis
[edit]The atoms that are used to build the purine nucleotides come from a variety of sources:
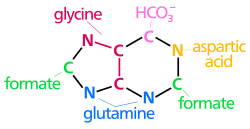
N1 arises from the amine group of Asp
C2 and C8 originate from formate
N3 and N9 are contributed by the amide group of Gln
C4, C5 and N7 are derived from Gly
C6 comes from HCO3− (CO2)

The de novo synthesis of purine nucleotides by which these precursors are incorporated into the purine ring proceeds by a 10-step pathway to the branch-point intermediate IMP, the nucleotide of the base hypoxanthine. AMP and GMP are subsequently synthesized from this intermediate via separate, two-step pathways. Thus, purine moieties are initially formed as part of the ribonucleotides rather than as free bases.
Six enzymes take part in IMP synthesis. Three of them are multifunctional:
The pathway starts with the formation of PRPP. PRPS1 is the enzyme that activates R5P, which is formed primarily by the pentose phosphate pathway, to PRPP by reacting it with ATP. The reaction is unusual in that a pyrophosphoryl group is directly transferred from ATP to C1 of R5P and that the product has the α configuration about C1. This reaction is also shared with the pathways for the synthesis of Trp, His, and the pyrimidine nucleotides. Being on a major metabolic crossroad and requiring much energy, this reaction is highly regulated.
In the first reaction unique to purine nucleotide biosynthesis, PPAT catalyzes the displacement of PRPP's pyrophosphate group (PPi) by an amide nitrogen donated from either glutamine (N), glycine (N&C), aspartate (N), folic acid (C1), or CO2. This is the committed step in purine synthesis. The reaction occurs with the inversion of configuration about ribose C1, thereby forming β-5-phosphorybosylamine (5-PRA) and establishing the anomeric form of the future nucleotide.
Next, a glycine is incorporated fueled by ATP hydrolysis, and the carboxyl group forms an amine bond to the NH2 previously introduced. A one-carbon unit from folic acid coenzyme N10-formyl-THF is then added to the amino group of the substituted glycine followed by the closure of the imidazole ring. Next, a second NH2 group is transferred from glutamine to the first carbon of the glycine unit. A carboxylation of the second carbon of the glycin unit is concomitantly added. This new carbon is modified by the addition of a third NH2 unit, this time transferred from an aspartate residue. Finally, a second one-carbon unit from formyl-THF is added to the nitrogen group and the ring is covalently closed to form the common purine precursor inosine monophosphate (IMP).
Inosine monophosphate is converted to adenosine monophosphate in two steps. First, GTP hydrolysis fuels the addition of aspartate to IMP by adenylosuccinate synthase, substituting the carbonyl oxygen for a nitrogen and forming the intermediate adenylosuccinate. Fumarate is then cleaved off forming adenosine monophosphate. This step is catalyzed by adenylosuccinate lyase.
Inosine monophosphate is converted to guanosine monophosphate by the oxidation of IMP forming xanthylate, followed by the insertion of an amino group at C2. NAD+ is the electron acceptor in the oxidation reaction. The amide group transfer from glutamine is fueled by ATP hydrolysis.
Pyrimidine and purine degradation
[edit]In humans, pyrimidine rings (C, T, U) can be degraded completely to CO2 and NH3 (urea excretion). That having been said, purine rings (G, A) cannot. Instead, they are degraded to the metabolically inert uric acid which is then excreted from the body. Uric acid is formed when GMP is split into the base guanine and ribose. Guanine is deaminated to xanthine which in turn is oxidized to uric acid. This last reaction is irreversible. Similarly, uric acid can be formed when AMP is deaminated to IMP from which the ribose unit is removed to form hypoxanthine. Hypoxanthine is oxidized to xanthine and finally to uric acid. Instead of uric acid secretion, guanine and IMP can be used for recycling purposes and nucleic acid synthesis in the presence of PRPP and aspartate (NH3 donor).[citation needed]
Prebiotic synthesis of nucleotides
[edit]Theories about the origin of life require knowledge of chemical pathways that permit formation of life's key building blocks under plausible prebiotic conditions. The RNA world hypothesis holds that in the primordial soup there existed free-floating ribonucleotides, the fundamental molecules that combine in series to form RNA. Complex molecules like RNA must have arisen from small molecules whose reactivity was governed by physico-chemical processes. RNA is composed of purine and pyrimidine nucleotides, both of which are necessary for reliable information transfer, and thus Darwinian evolution. Becker et al. showed how pyrimidine nucleosides can be synthesized from small molecules and ribose, driven solely by wet-dry cycles.[10] Purine nucleosides can be synthesized by a similar pathway. 5'-mono- and di-phosphates also form selectively from phosphate-containing minerals, allowing concurrent formation of polyribonucleotides with both the purine and pyrimidine bases. Thus a reaction network towards the purine and pyrimidine RNA building blocks can be established starting from simple atmospheric or volcanic molecules.[10]
Unnatural base pair (UBP)
[edit]An unnatural base pair (UBP) is a designed subunit (or nucleobase) of DNA which is created in a laboratory and does not occur in nature.[11] Examples include d5SICS and dNaM. These artificial nucleotides bearing hydrophobic nucleobases, feature two fused aromatic rings that form a (d5SICS–dNaM) complex or base pair in DNA.[12][13] E. coli have been induced to replicate a plasmid containing UBPs through multiple generations.[14] This is the first known example of a living organism passing along an expanded genetic code to subsequent generations.[12][15]
Medical applications of synthetic nucleotides
[edit]The applications of synthetic nucleotides vary widely and include disease diagnosis, treatment, or precision medicine.
- Antiviral or Antiretroviral agents: several nucleotide derivatives have been used in the treatment against infection with Hepatitis and HIV.[16][17] Examples of direct nucleoside analog reverse-transcriptase inhibitors (NRTIs) include Tenofovir disoproxil, Tenofovir alafenamide, and Sofosbuvir. On the other hand, agents such as Mericitabine, Lamivudine, Entecavir and Telbivudine must first undergo metabolization via phosphorylation to become activated.
- Antisense oligonucleotides (ASO): synthetic oligonucleotides have been used in the treatment of rare heritable diseases since they can bind specific RNA transcripts and ultimately modulate protein expression. Spinal muscular atrophy, amyotrophic lateral sclerosis, homozygous familial hypercholesterolemia, and primary hyperoxaluria type 1 are all amenable to ASO-based therapy.[18] The application of oligonucleotides is a new frontier in precision medicine and management of conditions which are untreatable.
- Synthetic guide RNA (gRNA): synthetic nucleotides can be used to design gRNA which are essential for the proper function of gene-editing technologies such as CRISPR-Cas9.
Length unit
[edit]Nucleotide (abbreviated "nt") is a common unit of length for single-stranded nucleic acids, similar to how base pair is a unit of length for double-stranded nucleic acids.[19]
Abbreviation codes for degenerate bases
[edit]The IUPAC has designated the symbols for nucleotides.[20] Apart from the five (A, G, C, T/U) bases, often degenerate bases are used especially for designing PCR primers. These nucleotide codes are listed here. Some primer sequences may also include the character "I", which codes for the non-standard nucleotide inosine. Inosine occurs in tRNAs and will pair with adenine, cytosine, or thymine. This character does not appear in the following table, however, because it does not represent a degeneracy. While inosine can serve a similar function as the degeneracy "D", it is an actual nucleotide, rather than a representation of a mix of nucleotides that covers each possible pairing needed.
| Symbol[20] | Description | Bases represented | ||||
|---|---|---|---|---|---|---|
| A | adenine | A | 1 | |||
| C | cytosine | C | ||||
| G | guanine | G | ||||
| T | thymine | T | ||||
| U | uracil | U | ||||
| W | weak | A | T | 2 | ||
| S | strong | C | G | |||
| M | amino | A | C | |||
| K | keto | G | T | |||
| R | purine | A | G | |||
| Y | pyrimidine | C | T | |||
| B | not A (B comes after A) | C | G | T | 3 | |
| D | not C (D comes after C) | A | G | T | ||
| H | not G (H comes after G) | A | C | T | ||
| V | not T (V comes after T and U) | A | C | G | ||
| N | any base (not a gap) | A | C | G | T | 4 |
See also
[edit]- Biology
- Chromosome
- Gene
- Genetics
- Nucleic acid analogue – Compound analogous to naturally occurring RNA and DNA
- Nucleic acid sequence – Succession of nucleotides in a nucleic acid
- Nucleobase – Nitrogen-containing biological compounds that form nucleosides
References
[edit]- ^ a b Zaharevitz DW, Anderson LW, Malinowski NM, Hyman R, Strong JM, Cysyk RL (November 1992). "Contribution of de-novo and salvage synthesis to the uracil nucleotide pool in mouse tissues and tumors in vivo". European Journal of Biochemistry. 210 (1): 293–6. doi:10.1111/j.1432-1033.1992.tb17420.x. PMID 1446677.
- ^ a b Alberts B, Johnson A, Lewis J, Raff M, Roberts K & Walter P (2002). Molecular Biology of the Cell (4th ed.). Garland Science. ISBN 0-8153-3218-1. pp. 120–121.
- ^ Abd El-Aleem FS, Taher MS, Lotfy SN, El-Massry KF, Fadel HH (2017-12-18). "Influence of extracted 5-nucleotides on aroma compounds and flavour acceptability of real beef soup". International Journal of Food Properties. 20 (sup1): S1182–S1194. doi:10.1080/10942912.2017.1286506. S2CID 100497537.
- ^ Wiley (2005-09-09). Encyclopedia of Life Sciences (1 ed.). Wiley. doi:10.1002/9780470015902.a0001333.pub3. ISBN 978-0-470-01617-6.
- ^ Smith AD, ed. (2000). Oxford Dictionary of Biochemistry and Molecular Biology (Revised ed.). Oxford: Oxford University Press. p. 460.
- ^ Ali E, Liponska A, O'Hara B, Amici D, Torno M, Gao P, et al. (June 2022). "The mTORC1-SLC4A7 axis stimulates bicarbonate import to enhance de novo nucleotide synthesis". Molecular Cell. 82 (1): 3284–3298.e7. doi:10.1016/j.molcel.2022.06.008. PMC 9444906. PMID 35772404.
- ^ See IUPAC nomenclature of organic chemistry for details on carbon residue numbering
- ^ Jones ME (1980). "Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis". Annual Review of Biochemistry. 49 (1): 253–79. doi:10.1146/annurev.bi.49.070180.001345. PMID 6105839.
- ^ McMurry JE, Begley TP (2005). The organic chemistry of biological pathways. Roberts & Company. ISBN 978-0-9747077-1-6.
- ^ a b Becker S, Feldmann J, Wiedemann S, Okamura H, Schneider C, Iwan K, et al. (October 2019). "Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides". Science. 366 (6461): 76–82. Bibcode:2019Sci...366...76B. doi:10.1126/science.aax2747. PMID 31604305. S2CID 203719976.
- ^ Malyshev DA, Dhami K, Quach HT, Lavergne T, Ordoukhanian P, Torkamani A, et al. (July 2012). "Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet". Proceedings of the National Academy of Sciences of the United States of America. 109 (30): 12005–10. Bibcode:2012PNAS..10912005M. doi:10.1073/pnas.1205176109. PMC 3409741. PMID 22773812.
- ^ a b Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, et al. (May 2014). "A semi-synthetic organism with an expanded genetic alphabet". Nature. 509 (7500): 385–8. Bibcode:2014Natur.509..385M. doi:10.1038/nature13314. PMC 4058825. PMID 24805238.
- ^ Callaway E (May 7, 2014). "Scientists Create First Living Organism With 'Artificial' DNA". Nature News. Huffington Post. Retrieved 8 May 2014.
- ^ Fikes BJ (May 8, 2014). "Life engineered with expanded genetic code". San Diego Union Tribune. Retrieved 8 May 2014.
- ^ Sample I (May 7, 2014). "First life forms to pass on artificial DNA engineered by US scientists". The Guardian. Retrieved 8 May 2014.
- ^ Ramesh D, Vijayakumar BG, Kannan T (December 2020). "Therapeutic potential of uracil and its derivatives in countering pathogenic and physiological disorders". European Journal of Medicinal Chemistry. 207: 112801. doi:10.1016/j.ejmech.2020.112801. PMID 32927231. S2CID 221724578.
- ^ Ramesh D, Vijayakumar BG, Kannan T (May 2021). "Advances in Nucleoside and Nucleotide Analogues in Tackling Human Immunodeficiency Virus and Hepatitis Virus Infections". ChemMedChem. 16 (9): 1403–1419. doi:10.1002/cmdc.202000849. PMID 33427377. S2CID 231576801. Archived from the original on 14 December 2021. Retrieved 13 March 2021.
- ^ Lauffer MC, van Roon-Mom W, Aartsma-Rus A (January 2024). "Possibilities and limitations of antisense oligonucleotide therapies for the treatment of monogenic disorders". Communications Medicine. 4 (1): 6. doi:10.1038/s43856-023-00419-1. PMC 10770028. PMID 38182878.
- ^ "Biology Terms Dictionary: nt". GenScript. Retrieved July 31, 2023.
- ^ a b Nomenclature Committee of the International Union of Biochemistry (NC-IUB) (1984). "Nomenclature for Incompletely Specified Bases in Nucleic Acid Sequences". Retrieved 2008-02-04.
Further reading
[edit]- Sigel A, Operschall BP, Sigel H (2017). "Chapter 11. Complex Formation of Lead(II) with Nucleotides and Their Constituents". In Astrid S, Helmut S, Sigel RK (eds.). Lead: Its Effects on Environment and Health. Metal Ions in Life Sciences. Vol. 17. de Gruyter. pp. 319–402. doi:10.1515/9783110434330-011. ISBN 9783110434330. PMID 28731304.
- Freisinger E, Sigel RK (July 2007). "From nucleotides to ribozymes—a comparison of their metal ion binding properties" (PDF). Coordination Chemistry Reviews. 251 (13–14): 1834–1851. doi:10.1016/j.ccr.2007.03.008.
- IUPAC-IUB Commission on Biochemical Nomenclature (CBN) (14 February 1971). "Abbreviations and Symbols for Nucleic Acids, Polynucleotides and their Constituents". Journal of Molecular Biology. 55 (3): 299–310. doi:10.1016/0022-2836(71)90319-6. PMID 5551389.
- Favre HA, Powell WH, eds. (2014). "Chapter P-10 Parent Structures for Natural Products and Related Compounds" (PDF). Nomenclature of Organic Chemistry: IUPAC Recommendations and Preferred Names 2013. Cambridge: Royal Soc. of Chemistry. ISBN 978-0-85404-182-4.
- Bender H, ed. (2003). "Nucleotide Structure". Clackamas Community College. Archived from the original on 2006-09-01. Retrieved 2020-04-21.